Classification and De-Identification of Free-Text Health Information via Machine Learning
by Ben Tefay, Senior Software Engineer
At Stacktrace, we believe that Machine Learning (ML) will be critical to improving healthcare delivery. One area that is ripe for improvement is automating the extraction of information from text documents, a task that has been intractable to software and expensive and laborious for humans. However, training an ML model to deliver this capability requires a large library of text documents, and these documents often contain sensitive identifying information about patients and health providers that can pose a substantial re-identification risk. Solving this problem of building a large dataset while respecting patient privacy and applicable local legislation is critical to training new ML models in a healthcare setting.
In this case study, we present our work on training an ML model to redact sensitive patient information in text documents. This model allows us to remove all direct patient identifiers and data representing a re-identification risk, such that we can safely and legally make use of these text documents to train downstream ML models. As an example, we then used these redacted documents to train a second machine learning system to classify text documents for further analysis.
The Importance of Machine Learning for Health Care
Personalised, evidence-based healthcare and research increasingly requires rich access to patient information. Much of this information, from patient histories to test results, is stored in unstructured text documents, which have historically been difficult for computers to understand. An inability to access this information has reduced the capability of software to support clinicians in providing improved health care. Areas such as automated auditing, decision support, diagnostic tools and large-scale analysis would be transformed by this capability. Excitingly, recent developments in Machine Learning (ML) have begun to enable software to deliver on this promise.
ML is revolutionary because it allows a computer program to learn from examples how to make predictions based on complex data. The resulting learnt software, called a model, can be far more complex than anything a human could build manually. To train a model, the software must be shown a large number of examples, where each example is an input - like a text document or image - that has been manually labelled with the desired prediction - like whether the text document contains a diagnosis of cancer. Creating the labelled examples to train an ML model requires significant effort by human experts. However, once the system is trained on these examples it can often outperform more traditional software and even approach expert competency in limited areas. The more labelled examples the model has seen, the better the model will perform (within limits).
Privacy and Machine Learning
One of the major impediments to using Machine Learning is the work required to build up a large collection of labelled examples to use for training. Collecting and using this dataset is particularly challenging in the case of medical documents because they contain sensitive patient information. In Australia, we are required by the Privacy Act to prevent sensitive patient information from being used for any purpose other than its stated intent. This typically does not include training an ML model, even if this model would eventually be used to improve their care. It is therefore important that any data used for training does not contain sensitive personal information that could be used to identify individuals.
To take the Privacy Act out of scope and make it legal to use sensitive medical information for ML, we need to transform the data such that it no longer qualifies as "sensitive personal information". To do this we need to:
- Remove all "direct identifiers". These are variables that can be used to uniquely identify an individual, either alone, or combined with other readily available information. The most common direct identifiers are names, addresses, dates of birth, hospital URNs and Medicare IHIs.
- Remove or otherwise alter the remaining information such that the risk of re-identification is minimized. This is tricky, because the risk of re-identification depends on who has access to the data, and the other data they may have. For example, a record that is going to be published on a website could be read by anyone, and so must be extensively redacted to ensure there is a low risk the patient can be reidentified. A record that is only accessible to a pathology lab, on the other hand, can contain far more information without significant risk of the patient being reidentified.
De-identification is a nuanced problem. The goal is to preserve your patient's privacy, while still allowing you to derive the insights you need. It requires data custodians to balance the data's utility and the level of re-identification risk which remains after de-identification. This risk is highly context specific, depending on how likely a patient's information can be tied back to the patient.
For more information, we have found the following links helpful:
- An introduction to de-identification: De-identification and the Privacy Act
- Pragmatic advice: A Framework for Data De-identification
- Further reading: Australian Privacy Principles and The Privacy Act
Training a Machine Learning System to De-Identify Patient Data
In order to train and use ML models to extract information from text-based health documents, we first require a large number of examples of the documents we would like to analyse. To legally store and use these documents, we must ensure they do not contain sensitive patient information. Manually redacting this patient information would be both impractical and prohibitively expensive for the volumes of patients seen by even a single clinic. To address this problem, we set out to train an ML model that can automatically remove sensitive patient information from documents.
As a first step, we created a dataset of examples by manually labelling all of the sensitive patient and medical practioner information in a collection of textual pathology reports and letters. This involved reading each documented and highlighting instances of sensitive information, including names, businesses, addresses, dates and times (if more specific than a month), identifying numbers and titles. On average, these documents contained 10 instances of personally identifiable information (PII) about doctors and patients per document.
Using the spaCy natural language processing library, we trained a named entity recognizer (a type of ML model) on 80% of these labelled documents. An example of the output from the trained model when run over a synthetic text document is shown in Figure 1.
We then evaluated the model's performance by running the algorithm over the remaining 20% of the documents (which it had never seen before), and compared its predictions to our own labels.
The results were as follows:
- It correctly redacted 97.23% of the PII in the documents. This means that 2.77% of the PII in the documents was missed by the model and not redacted, which is a potential re-identification risk.
- Of all the spans of text it redacted, 97.68% was actually PII. This means that 2.32% of the redacted spans of text were incorrectly marked as PII, and did not need to be redacted. This is data that we have unnecessarily lost, and which otherwise could have been useful.
Combining these two results into an F1 score, which is a metric typically used in the ML field, the model achieved an overall performance of 97.45%.
While the model is not perfect, it is a positive indicator that de-identification can be automated for this type of document. The model could be included in a pipeline that, combined with human oversight, automatically pulled text documents from a practice management or electronic medical record system for further analysis downstream. Furthermore, the model could be updated over time as it received feedback from human auditors, allowing its performance to improve even further.

Figure 1. An example of the output from our PII model when run over synthetic data. All of the spans of text it has predicted as PII have been marked in yellow. We consider names, businesses, addresses, dates and times (if more specific than a month), identifying numbers and titles to be sensitive information. In this case, the model has correctly found all of the PII. However, it has also incorrectly labelled the medication “Mini Pill” and obstetric history “G0P0” as PII.
Using the De-Identified Text Documents to Train a Document Classifier
As a proof of concept that we could use the redacted output from our de-identification model to train downstream ML models, we trained a second model to classify documents into one of 5 classes, including histology, cytology, blood test and imaging results, and letters. An example of the output from the model is shown in Figure 2.
The classifier achieved an F1 score of 94.88% (a typical indicator of accuracy in the ML field). This far outperforms our existing software solution, and is strong evidence that our approach to de-identification and ML is viable.
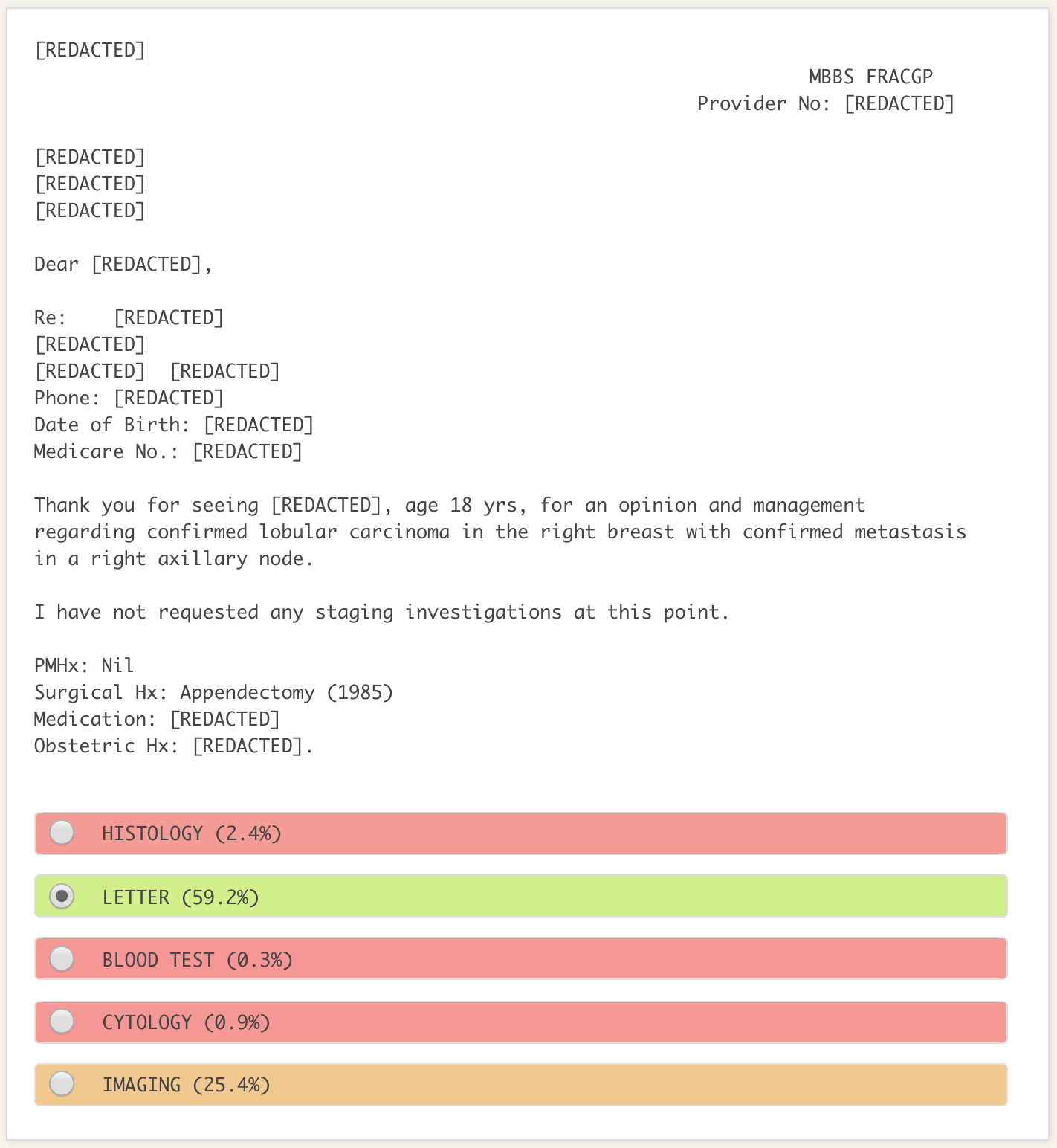
Figure 2. An example of the output from our pathology result classifier when run over synthetic data. The model has 59% confidence that this is a letter, 25% confidence this is an imaging result, and so on. The model will take the highest confidence category as its prediction, so in this case it will correctly predict that this is a letter. While the model believes there is a 25% chance this is an imaging result, we do not know why. Being unable to explain the reasoning of the model is a common problem in ML.
Let’s Work Together
We have made exciting progress on using ML to de-identify and extract useful information from text documents. However, this is just the beginning.
Our models are only as good as the data they have been trained on - and ours have only been exposed to a specific set of pathology reports and letters. This means that their utility is likely limited to text documents that have a similar structure and content. Much more useful are the workflows and skillset that we have built for importing and labelling data and training ML models.
We are excited to understand how we can work with you to apply these skills and tools to your specific problems. Whether this is building a bespoke ML solution for your text processing requirements, or extending our existing models to work with your dataset, we would love to help.